tcrdist2¶
2020-06-27
tcrdist2 provides flexible distance measures for comparing T cell receptors across complementarity determining regions (CDRs)
tcrdist2 is a python API-enabled toolkit for analyzing T-cell receptor repertoires. Some of the functionality and code is adapted from the original tcr-dist package which was released with the publication of Dash et al. Nature (2017). The manuscript (doi:10.1038/nature22383). This package contains a new API for accessing the features of tcr-dist, as well as many new features that expand the T cell receptor analysis pipeline.
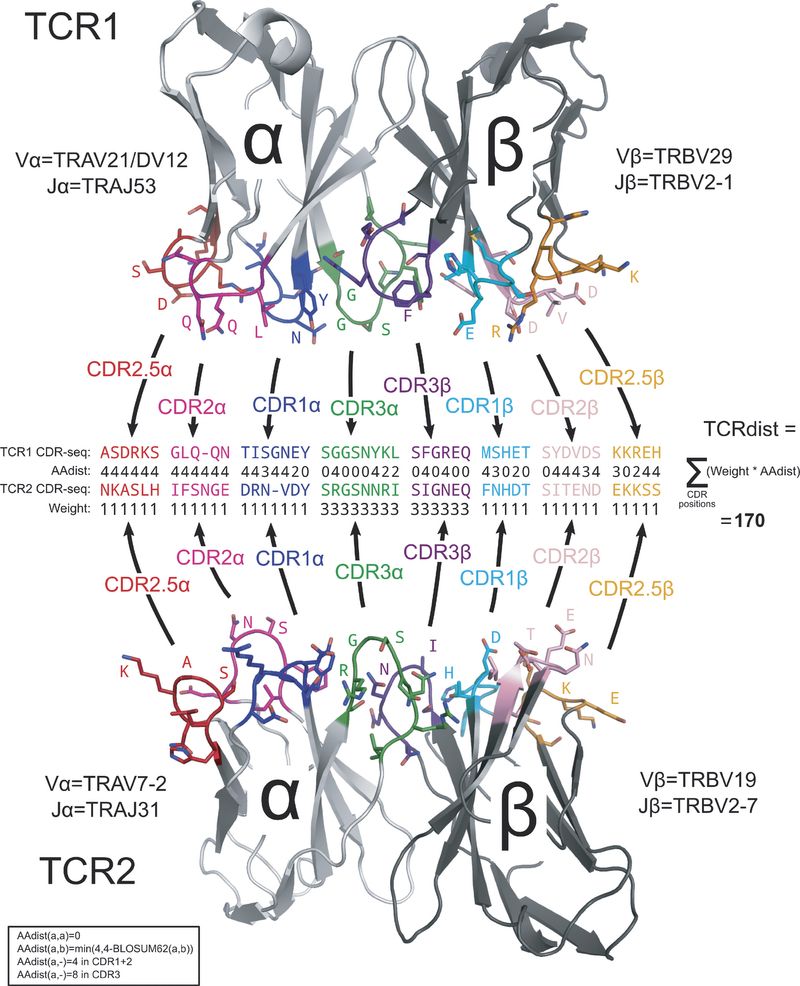
Extended Data Figure 3 : Schematic overview of the TCRdist calculation.
Extended Data Figure 3 is particularly illustrative.
“Each TCR is mapped to the amino acid sequences of the loops within the receptor that are known to provide contacts to the pMHC (commonly referred to as CDR1, CDR2, and CDR3, as well as an additional variable loop between CDR2 and CDR3). The distance between two TCRs is computed by comparing these concatenated CDR sequences using a similarity-weighted Hamming distance, with a gap penalty introduced to capture variation in length and a higher weight given to the CDR3 sequence in recognition of its disproportionate role in epitope specificity (see Methods and Extended Data Fig. 3).”
Getting started¶
Quick Examples¶
Long Examples¶
Pre-Processing¶
In-Depth¶